Understanding Support Vector Machines: A Beginner’s Guide

Support Vector Machines (SVM) are powerful supervised learning algorithms used for classification and regression tasks. In this guide, we’ll break down how SVMs work, why they’re useful, and how to implement them in Python. Let’s start from the basics and work our way up to practical applications.
1. What is a Support Vector Machine?
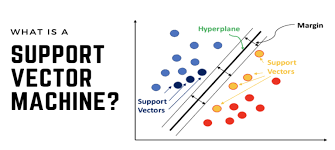
Think of SVM as a method that tries to find the best dividing line (or hyperplane in higher dimensions) between different groups of data points. What makes it special is that it doesn’t just find any dividing line – it finds the optimal one by maximizing the margin between the closest points of different classes.
Key Concepts:
- Hyperplane: The decision boundary that separates different classes
- Support Vectors: The data points closest to the hyperplane
- Margin: The distance between the support vectors and the hyperplane
2. How Does SVM Work?
Linear SVM: The Simple Case
Imagine you have two groups of points on a plane that can be separated by a straight line. SVM will:
- Find multiple possible lines that could separate the data
- Calculate the distance (margin) from each line to the nearest points
- Choose the line that has the largest margin
Non-linear SVM: The Kernel Trick
But what if your data isn’t linearly separable? This is where the “kernel trick” comes in. SVM can transform the data into a higher dimension where it becomes linearly separable.
3. Practical Implementation
Let’s implement SVM using Python and scikit-learn. We’ll start with a simple example:
import numpy as np
from sklearn import datasets, svm
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
# Generate sample data
X, y = datasets.make_blobs(n_samples=100, centers=2, random_state=42)
# Split the data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Create and train the SVM classifier
clf = svm.SVC(kernel='linear')
clf.fit(X_train, y_train)
# Make predictions
predictions = clf.predict(X_test)
# Plot the results
def plot_svm_boundary(X, y, clf):
plt.scatter(X[:, 0], X[:, 1], c=y, cmap='rainbow')
# Create a mesh grid
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
# Plot decision boundary
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z)
plt.show()
plot_svm_boundary(X, y, clf)4. Important SVM Parameters
Kernel Types
- Linear:
kernel='linear'- Best for linearly separable data
- RBF (Radial Basis Function):
kernel='rbf'- Good for non-linear data
- Polynomial:
kernel='poly'- Useful for curved decision boundaries
Let’s see how different kernels perform on non-linear data:
# Generate non-linear data
X, y = datasets.make_moons(n_samples=100, noise=0.15)
# Create classifiers with different kernels
classifiers = {
'Linear SVM': svm.SVC(kernel='linear'),
'RBF SVM': svm.SVC(kernel='rbf'),
'Polynomial SVM': svm.SVC(kernel='poly', degree=3)
}
# Train and plot each classifier
for name, clf in classifiers.items():
clf.fit(X, y)
plt.figure(figsize=(8, 6))
plt.title(name)
plot_svm_boundary(X, y, clf)5. Real-World Example: Iris Classification
Let’s apply SVM to the famous Iris dataset:
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score, classification_report
# Load iris dataset
iris = load_iris()
X = iris.data
y = iris.target
# Split the data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
# Create and train the model
clf = svm.SVC(kernel='rbf', C=1.0)
clf.fit(X_train, y_train)
# Make predictions
y_pred = clf.predict(X_test)
# Print results
print("Accuracy:", accuracy_score(y_test, y_pred))
print("nClassification Report:n", classification_report(y_test, y_pred))6. Best Practices and Tips
Data Preprocessing
- Always scale your features (use StandardScaler or MinMaxScaler)
- Handle missing values
- Convert categorical variables
Parameter Tuning
- Use GridSearchCV for finding optimal parameters
- Key parameters to tune:
- C (regularization parameter)
- kernel
- gamma (for RBF kernel)
from sklearn.model_selection import GridSearchCV
# Define parameter grid
param_grid = {
'C': [0.1, 1, 10],
'kernel': ['rbf', 'linear'],
'gamma': ['scale', 'auto', 0.1, 1],
}
# Create GridSearchCV object
grid_search = GridSearchCV(svm.SVC(), param_grid, cv=5)
grid_search.fit(X_train, y_train)
print("Best parameters:", grid_search.best_params_)
print("Best score:", grid_search.best_score_)7. When to Use SVM?
SVMs are particularly good for:
- Medium-sized datasets
- High-dimensional spaces
- Cases where you need a clear margin of separation
- Both linear and non-linear classification
However, they might not be the best choice for:
- Very large datasets (can be computationally expensive)
- Lots of noise in the data
- Cases where you need probability estimates
What’s Next ?
Support Vector Machines are versatile algorithms that can handle both linear and non-linear classification tasks. By understanding the basic concepts and following the implementation guidelines above, you can effectively use SVMs in your machine learning projects.
Remember to:
- Start with simple linear kernels
- Scale your data
- Use cross-validation for parameter tuning
- Consider computational resources for larger datasets
Practice with the code examples provided, and gradually work your way up to more complex applications. Happy learning!
In-case you have faced any difficult, please make a good use of the comment section i will personal be there to help you when you are stuck.
We think sharing practical implementation on real world example various machine learning skill is the key point to mastery and also solve various problem that affect our society we are intended to teach you through practical means if you think our idea is good. Please and please leave us a comment below about your views or request an article. as usual don’t forget to up-vote this article and share it.



